論文発表
31. Loss of non-canonical translation initiation factors impairs perinatal cardiac function in mice
Asai T, Tochinai R, Tsuru Y, Sekiguchi M, Minami A, Fujii W, Kyuwa S, Ogawa T, Kakuta S*.
Exp Anim. 2025 Oct 8. doi: 10.1538/expanim.25-0021
30. The ribonuclease RNase T2 mediates selective autophagy of ribosomes induced by starvation in Saccharomyces cerevisiae
Minami A, Nishi K, Yamada R, Jinnai G, Shima H, Oishi S, Akagawa H, Aono T, Hidaka M, Masaki H, Kuzuyama T, Noda Y, Ogawa T*.
J. Biol. Chem. 2025 Apr 26. doi: 10.1016/j.jbc.2025.108554
29. Mechanistic Characterization of Diterpene Synthase Pairs for Tricyclic Diterpenes from Cyanobacteria.
Yu J, Shiraishi T, Taizoumbe KA, Karasuno Y, Yoshida A, Nishiyama M, Dickschat JS*, Kuzuyama T*.
J Am Chem Soc. 2025 Mar 27. doi: 10.1021/jacs.4c16710.
28. IER5 Promotes Ovarian Cancer Cell Proliferation and Peritoneal Dissemination.
Krishnaraj J, Ueno S, Nakamura M, Tabata Y, Yamamoto T, Asano Y, Tanaka T, Kuzuyama T, Saya H, Ohki R*.
Cancers (Basel). 2025 doi: 10.3390/cancers17040610
27. In vivo and in vitro reconstitution of biosynthesis of N-prenylated phenazines revealing diverse phenazine-modifying enzymes.
Kato T, Xia D, Ozaki T, Nakao T, Zhao P, Nishiyama M, Shiraishi T, Kuzuyama T*.
ChemBioChem. 2024 doi: 10.1002/cbic.202400723.
26. ‘Tuning’ of ribosome levels mediated by RNase I and hibernating ribosomes.
Minami A, Tanzawa T*, Yang Z, Funatsu T, Kuzuyama T, Yoshida H, Kato T, Ogawa T*.
bioRxiv. 2024 doi: 10.1101/2024.07.29.605612.
25. Establishment of the improved colonization of Escherichia coli laboratory strain in the intestine mediated by single gene deletion.
Minami A, Asai T, Tachibana T, Tanaka Y, Nakajima M, Tamura S, Nakazawa M, Tsuru Y, Fujiyama Y, Tagawa YI, Kuzuyama T, Kakuta S*, Ogawa T*.
Biochem Biophys Res Commun. 2024 Nov 19;734:150448. doi: 10.1016/j.bbrc.2024.150448.
24. QM/MM Study of the Catalytic Mechanism and Substrate Specificity of the Aromatic Substrate C-Methyltransferase Fur6.
Zhao F, Moriwaki Y, Noguchi T, Shimizu K, Kuzuyama T, Terada T*.
Biochemistry. 2024 Mar 19;63(6):806. doi: 10.1021/acs.biochem.3c00556.
23. RNase T2-involved selective autophagy of ribosomes induced by starvation in yeast.
Minami A, Nishi K, Yamada R, Jinnai G, Shima H, Oishi S, Akagawa H, Aono T, Hidaka M, Masaki H, Kuzuyama T, Noda Y, Ogawa T*.
bioRxiv. 2023 doi: 10.1101/2023.07.14.548783.
22. Discovery of type II polyketide synthase-like enzymes for the biosynthesis of cispentacin.
Hibi G, Shiraishi T, Umemura T, Nemoto K, Ogura Y, Nishiyama M, Kuzuyama T*.
Nat Commun. 2023 Dec 6;14(1):8065. doi: 10.1038/s41467-023-43731-z.
press release
21. Dethiobiotin uptake and utilization by bacteria possessing bioYB operon.
Ikeda T, Ogawa T, Aono T*.
Res Microbiol. 2023 Nov-Dec;174(8):104131. doi: 10.1016/j.resmic.2023.104131.
20. Substrate Recognition Mechanism of a Trichostatin A-Forming Hydroxyamidotransferase.
Nagata R*, Nishiyama M, Kuzuyama T*.
Biochemistry 2023 Jun 20;62(12):1833-1837. doi: 10.1021/acs.biochem.3c00025.
Selected as Front Cover Art!
19. Complete Biosynthetic Pathway of the Phosphonate Phosphonothrixin: Two Distinct Thiamine Diphosphate-Dependent Enzymes Divide the Work to Form a C–C Bond.
Zhu Y, Shiraishi T, Lin J, Inaba K, Ito A, Ogura Y, Nishiyama M, Kuzuyama T*.
J Am Chem Soc. 2022 Sep 21;144(37):16715-16719. doi: 10.1021/jacs.2c06546.
This article was introduced in “Hot of the press”(Natural Product Reports)
18. Cryptic Oxidative Transamination of Hydroxynaphthoquinone in Natural Product Biosynthesis.
Noguchi T, Isogai S, Terada T, Nishiyama M, Kuzuyama T*.
J Am Chem Soc. 2022 Mar 30;144(12):5435-5440. doi: 10.1021/jacs.1c13074.
press release
17. Structural Basis for the Prenylation Reaction of Carbazole-Containing Natural Products Catalyzed by Squalene Synthase-like Enzymes.
Nagata R, Suemune H, Kobayashi M, Shinada T, Shin-ya K, Nishiyama M, Hino T, Sato Y, Kuzuyama T*, Nagano S*.
Angew Chem Int Ed Engl. 2022 Mar 9:61(20):e202117430. doi: 10.1002/anie.202117430.
press release
16. Total Synthesis and Stereochemistry Assignment of Nucleoside Antibiotic A-94964.
Shao X, Zheng C, Xu P, Shiraishi T, Kuzuyama T, Molinaro A, Silipo A, Yu B.
Angew Chem Int Ed Engl. 2022 Mar 28;61(14):e202200818. doi: 10.1002/anie.202200818.
15. Biosynthesis of the nucleoside antibiotic angustmycins: identification and characterization of the biosynthetic gene cluster reveal unprecedented dehydratase required for exo-glycal formation.
Shiraishi T, Xia J, Kato T, Kuzuyama T.*
J Antibiot. 2021 Nov;74(11):830-833. doi: 10.1038/s41429-021-00466-7.
14. A novel oxazole-containing tetraene compound, JBIR-159, produced by heterologous expression of the cryptic trans-AT type polyketide synthase biosynthetic gene cluster.
Hashimoto T, Hashimoto J, Kagaya N, Nishimura T, Suenaga H, Nishiyama M, Kuzuyama T, Shin-ya K.*
J Antibiot. 2021 May;74(5):354-358. doi: 10.1038/s41429-021-00410-9.
13. Biosynthetic pathways and enzymes involved in the production of phosphonic acid natural products.
Shiraishi T, Kuzuyama T.*
Biosci Biotechnol Biochem. 2021 Jan 7;85(1):42-52. doi: 10.1093/bbb/zbaa052.
Selected as a cover picture!
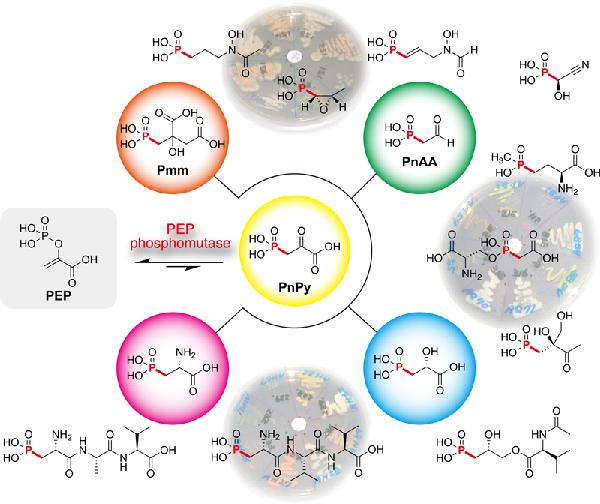
12. Reconstitution of a Highly Reducing Type II PKS System Reveals 6π-Electrocyclization Is Required for o-Dialkylbenzene Biosynthesis.
Zhang J, Yuzawa S, Thong WL, Shinada T, Nishiyama M, Kuzuyama T.*
J Am Chem Soc. 2021 Fub 24;143(7):2962-2969. doi: 10.1021/jacs.0c13378.
press release
11. Substrate recognition mechanism of tRNA-targeting ribonuclease, colicin D, and an insight into tRNA cleavage-mediated translation impairment.
Ogawa T, Takahashi K, Ishida W, Aono T, Hidaka M, Terada T, Masaki H.*
RNA Biol. 2021 Aug;18(8):1193-1205. doi: 10.1080/15476286.2020.1838782.
press release

10. Recent advances in the biosynthesis of nucleoside antibiotics.
Shiraishi T, Kuzuyama T.*
J Antibiot. 2019 Dec;72(12):913-923. doi: 10.1038/s41429-019-0236-2.
9. An Unusual Skeletal Rearrangement in the Biosynthesis of the Sesquiterpene Trichobrasilenol from Trichoderma.
Murai K,† Lauterbach L,† Teramoto K, Quan Z, Barra L, Yamamoto T, Nonaka K, Shiomi K, Nishiyama M, Kuzuyama T,* Dickschat JS.*
Angew Chem Int Ed Engl. 2019 Aug 16. doi: 10.1002/anie.201907964.

8. An Unprecedented Cyclization Mechanism in the Biosynthesis of Carbazole Alkaloids in Streptomyces.
Kobayashi M, Tomita T, Shin-ya K, Nishiyama M, Kuzuyama T.*
Angew Chem Int Ed Engl. 2019 Jul 26. doi: 10.1002/anie.201906864.
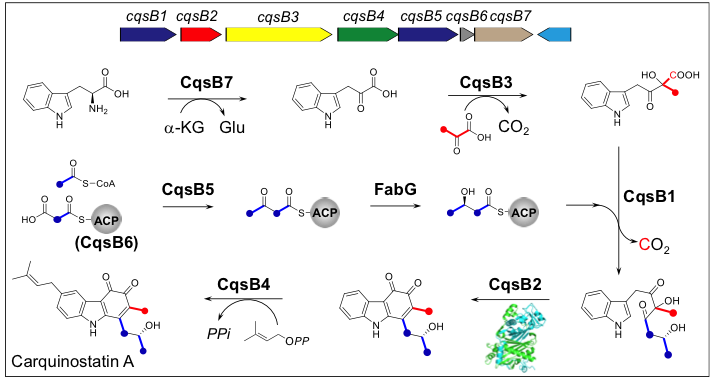
7. Biochemical characterization of archaeal homocitrate synthase from Sulfolobus acidocaldarius.
Suzuki T, Akiyama N, Yoshida A, Tomita T, Lassak K, Haurat MF, Okada T, Takahashi K, Albers SV, Kuzuyama T, Nishiyama M.*
FEBS Lett. 2019 Jul 22. doi: 10.1002/1873-3468.13550.
6. Comprehensive Derivatization of Thioviridamides by Heterologous Expression.
Kudo K, Koiwai H, Kagaya N, Nishiyama M, Kuzuyama T, Shin-ya K, Ikeda H.*
ACS Chem Biol. 2019 Jun 21;14(6):1135-1140. doi: 10.1021/acschembio.9b00330.
5. The Amipurimycin and Miharamycin Biosynthetic Gene Clusters: Unraveling the Origins of 2-Aminopurinyl Peptidyl Nucleoside Antibiotics.
Romo AJ,† Shiraishi T,† Ikeuchi H, Lin GM, Geng Y, Lee YH, Liem PH, Ma T, Ogasawara Y, Shin-ya K, Nishiyama M, Kuzuyama T,* Liu HW.*
J Am Chem Soc. 2019 May 31. doi: 10.1021/jacs.9b03021.
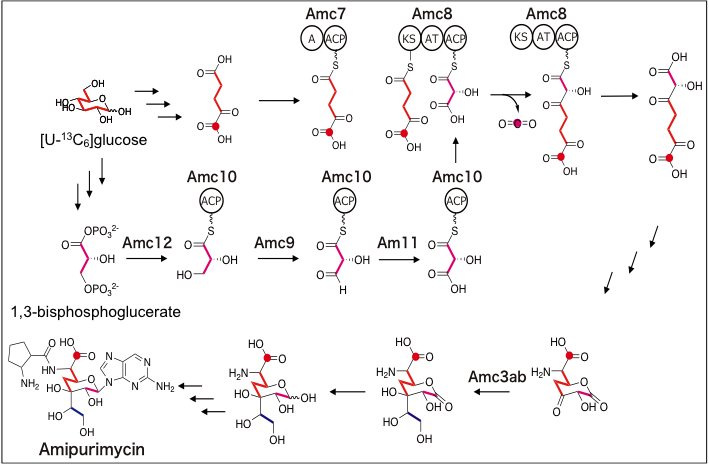
4. Glutamate Dehydrogenase from Thermus thermophilus Is Activated by AMP and Leucine as a Complex with Catalytically Inactive Adenine Phosphoribosyltransferase Homolog.
Tomita T, Matsushita H, Yoshida A, Kosono S, Yoshida M, Kuzuyama T, Nishiyama M.*
J Bacteriol. 2019 Jun 21;201(14). pii: e00710-18. doi: 10.1128/JB.00710-18.
3. Protein acetylation on 2-isopropylmalate synthase from Thermus thermophilus HB27.
Yoshida A, Yoshida M, Kuzuyama T, Nishiyama M, Kosono S.*
Extremophiles. 2019 Jul;23(4):377-388. doi: 10.1007/s00792-019-01090-y.
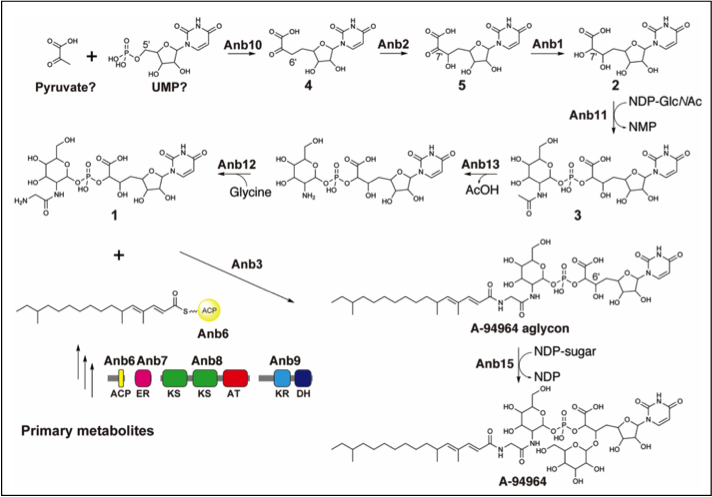
2. Biosynthesis of the uridine-derived nucleoside antibiotic A-94964: identification and characterization of the biosynthetic gene cluster provide insight into the biosynthetic pathway.
Shiraishi T, Nishiyama M, Kuzuyama T.*
Org Biomol Chem. 2019 Jan 16;17(3):461-466. doi: 10.1039/c8ob02765j.
1. Structural and Mechanistic Insight into Terpene Synthases that Catalyze the Irregular Non-Head-to-Tail Coupling of Prenyl Substrates.
Kobayashi M, Kuzuyama T.*
Chembiochem. 2019 Jan 2;20(1):29-33. doi: 10.1002/cbic.201800510.
学会等での受賞
- 2024/03/22
倉本 泰遥(B4) 令和5年度東京大学農学部 学部長賞 - 2024/03/21
日比 玄紀(D3) 令和5年度東京大学大学院農学生命科学研究科 研究科長賞 - 2023/09/29
尉 嘉眙(D2) 第33回イソプレノイド研究会例会 奨励賞 - 2023/09/07
白石 太郎(助教) 2023年度日本放線菌学会 浜田賞 - 2023/07/07
南 篤(D3) 第24回日本RNA学会年会 年会長特別賞 - 2023/03/14
朱 裕勋(D2) 日本農芸化学会2023年度大会 トピックス賞 - 2022/09/29
南 篤(D2) 第60回日本生物物理学会年会 シンポジウム演題採択 - 2022/09/16
朱 裕勋(D2) 2022年度日本放線菌学会大会 ポスター発表賞 - 2022/08/27
日比 玄紀(D2) 日本農芸化学会関東支部2022年度大会 優秀発表賞 - 2022/06/30
朱 裕勋(D2) 4th Global Innovation Workshop Outstanding Presentation Award
徐 昊(M2) 4th Global Innovation Workshop Poster Award - 2022/03/17
日比 玄紀(D1) 日本農芸化学会2022年度大会 トピックス賞 - 2021/11/05
野口 智弘(D3) 第63回天然有機化合物討論会 奨励賞 - 2021/09/18
湯澤 賢(共同研究員) 2021年度日本放線菌学会 浜田賞 - 2021/08/28
野口 智弘(D3) 2021年度日本農芸化学会関東支部大会 優秀発表賞 - 2020/11/10
南 篤(M2) 2nd Global Innovation Workshop Outstanding Presentation Award - 2020/10/30
吉留 大輔(D3) 第20回東京大学生命科学シンポジウム 優秀ポスター賞
